Paper II
Genetic overlap between multivariate measures of human functional brain connectivity and psychiatric disorders
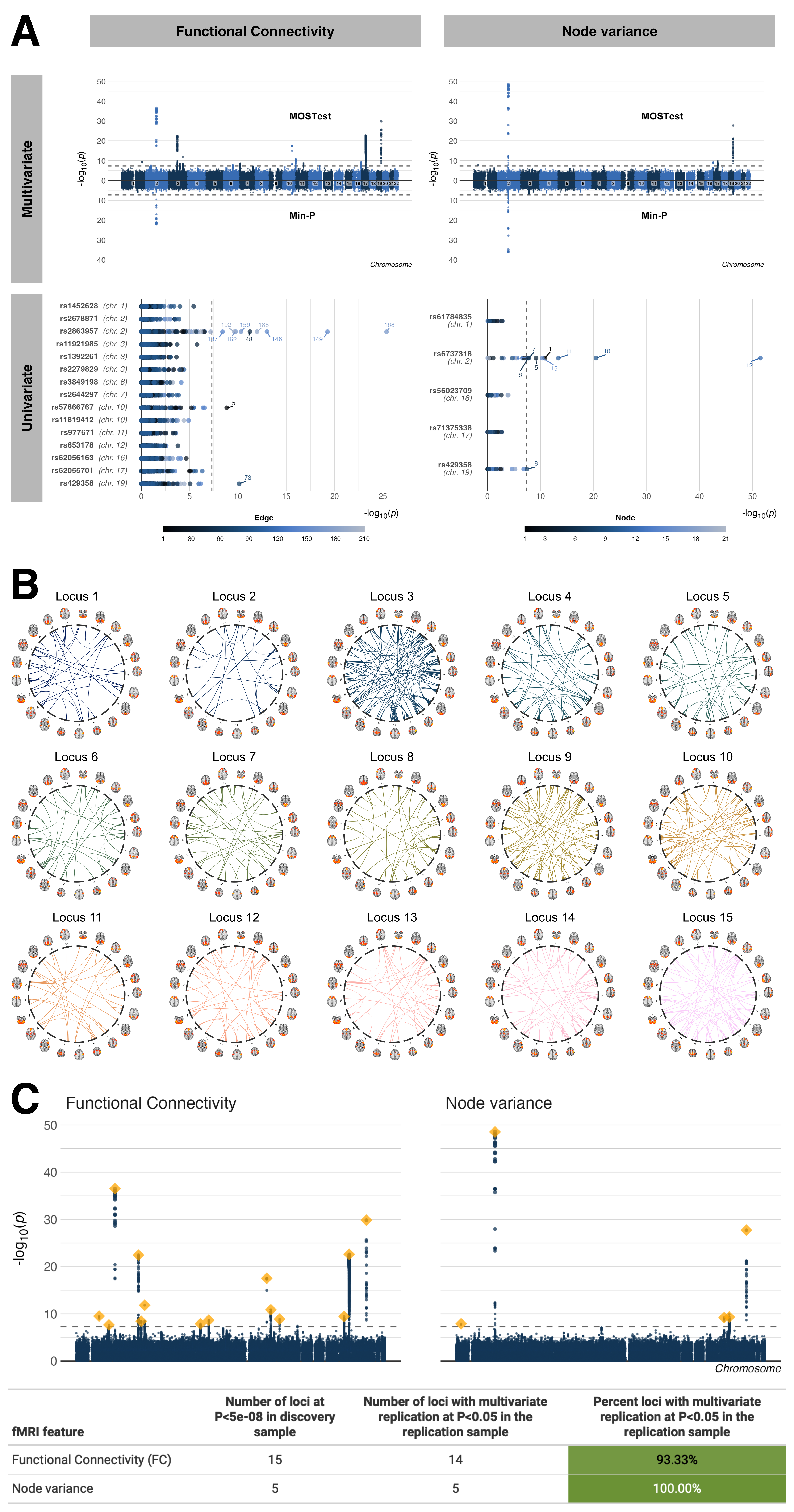
We applied a multivariate GWAS framework implemented through MOSTest1 to fMRI features on FC (210 features) and temporal node variance (21 features) from 30,701 individuals in the UKB. We identified 15 independent genetic loci significantly associated (p < 5e-8) with FC and 5 loci associated with temporal node variance through the multivariate analysis. The univariate stream identified 2 and 3 independent loci respectively. In an independent replication sample nearly all of these loci replicated at nominal p-value (14/15, 93%, p < 0.05). Figure 26.1 panel A shows the Miami plots for both features for both the multivariate and univariate stream. Figure 26.1 panel B shows the distributed nature of these loci across the brain, where a single locus is significantly (p < 0.05) associated with a number of univariate comparisons.
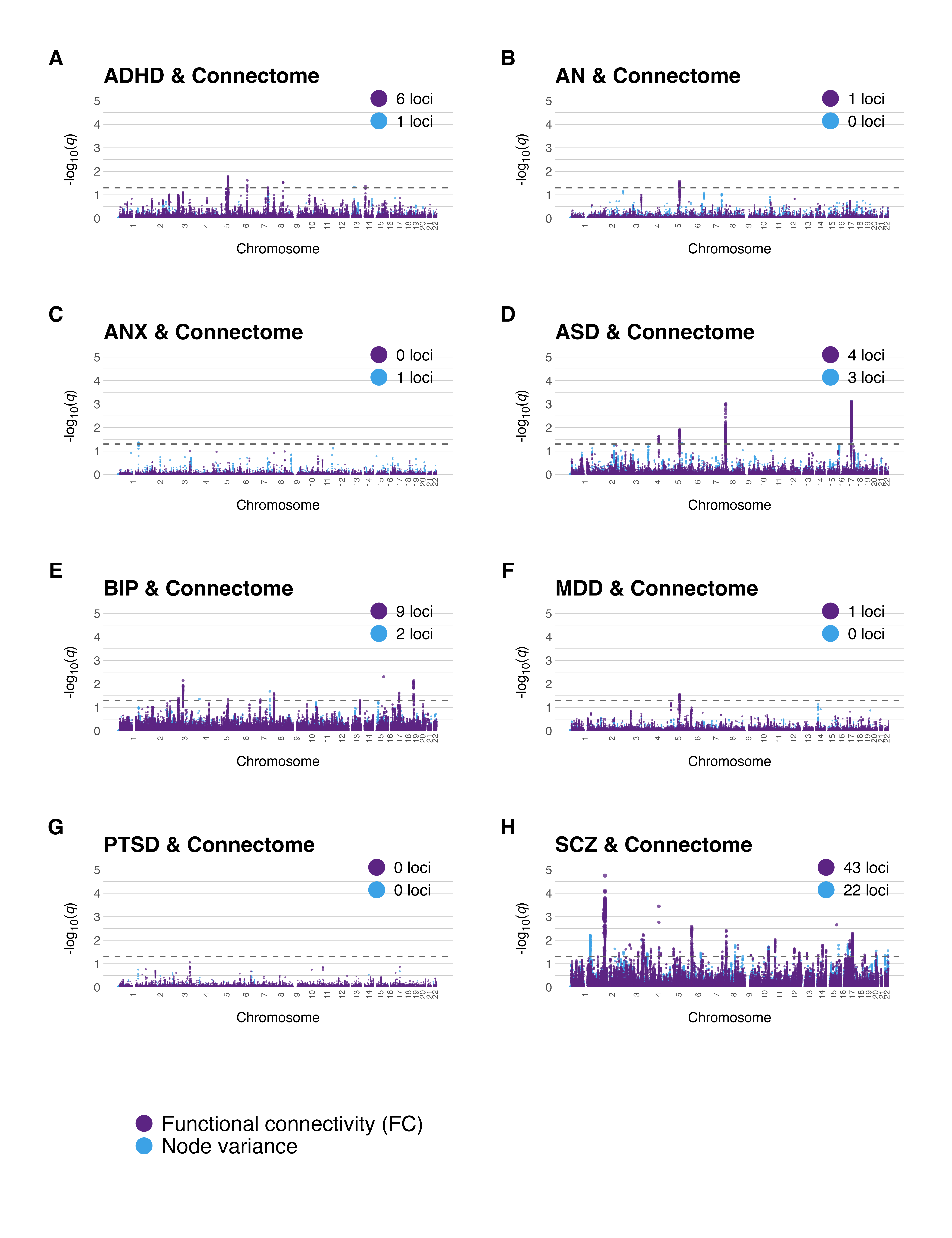
Next we compared the MOSTest summary statistics on FC and node variance with seven psychiatric disorders (ADHD, ASD, ANX, BIP, MD, PTSD, and SCZ) using conjFDR analysis. As indicated in Figure 26.2, we found overlapping loci with the brain connectivity features for six out of the seven disorders in this analysis. We found the largest overlap with SCZ, 43 overlapping loci for FC and 22 for node variance. Comparisons with a negative control trait, in this case vitamin D levels (N = 79,366) showed 2 overlapping loci with FC and none with node variance.
Using FUMA3 we mapped the shared loci between the imaging features and the psychiatric conditions to the genes associated with these loci. This yielded 180 genes associated with any of the features or disorders in our analysis. Next, we analyzed these genes with the SynGO4 tool to identify associations of these genes with synaptic processes. We found associations with synaptic functioning for 23 out of 180 genes, such as bdnf, a major regulator of synaptic transmission and synaptic plasticity5, and nrxn1, which plays a role in the formation of synapstic connections6. The ability to implicate these synapse-related genes provides further support from an imaging-genetics perspective for the dysconnectivity hypothesis of psychiatric disorders, as discussed more closely in the Discussion ?sec-discussion section of this thesis.